Note
Click here to download the full example code
Variable Pacing Frequency¶
This example illustrates how to setup a simulation which modifies the protocol while it is running. In this case we will show how this can be used to change the pacing frequency (basic cycle length), however in general this can be applied to change any parameter during runtime.
Initial Setup¶
First we will setup paced current clamp simulation with any of the options we would like.
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
import PyLongQt as pylqt
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
proto = pylqt.protoMap['Current Clamp Protocol']()
proto.setCellByName('Canine Ventricular (Hund-Rudy 2009)')
proto.bcl = 2_000
proto.tMax = 55_000
proto.numstims = 200
proto.meastime = 1_000
proto.writetime = 0
proto.measureMgr.addMeasure('vOld', {'cl', 'peak'})
Now that the basic setup is complete we will use an additional feature
of Protocol, Protocol.setRunDuring(), which allows
us to set a function to be called periodically while the simulation is
running. The settings for the run durring function are the first time
for the function to be called firstRun (in ms), how often it should be run
runEvery (in ms) and finally how many times it should be run: numruns.
Note
There are two other features which similarly allow for a user defined function to be run during the simulation: setRunBefore and setRunAfter. As the names imply these will be run at the beginning and end of a simulation.
Define the run during function¶
The user defined function we will supply to the method will be a special one. In python it is possible to define a class which can act like a function, and we will do that in order to allow our ‘function’ to store all of the `bcl`s.
To do this we will define the class IterateBCLs, which will take the list of bcls we want to use, and then update the protocol every time it is called as a function.
class IterateBCLs():
def __init__(self, bcls):
self.proto_lookup = {}
self.bcls = bcls
def __len__(self):
return len(self.bcls)
def __call__(self, proto):
if not id(proto) in self.proto_lookup:
self.proto_lookup[id(proto)] = 0
idx = self.proto_lookup[id(proto)]
if idx < len(self.bcls):
bcl = self.bcls[idx]
proto.bcl = bcl
self.proto_lookup[id(proto)] += 1
else:
proto.paceflag = False
A detailed explanation for those unfamiliar with python classes
The __init__ method is the one that is called when IterateBCLs is created. In our case it initializes the proto_lookup table, and saves the bcls for use later. To call this method run
func = IterateBCLs([1000,500]). The __len__ function returns the number of bcls that are stored, and could be calledlen(func). Finally, the most important method is __call__, this is what will be called while the simulation is running. It can be called usingfunc(proto).
The __call__ method preforms the following tasks:
Determines the number of times that the __call__ method has been called so that it will know which bcl to assign to the protocol
If the number of times it has been called is less than the number of bcls passed in, it updates the protocol’s bcl to the next bcl in the list, otherwise it instructs the protocol to stop pacing.
For step 1, we need to use a lookup table for the case where we are running multiple simulations using numtrials. That is not the case in this example, but this allows the IterateBCLs class to be more generally useful. The lookup table is needed to ensure that IterateBCLs keeps track of the number of calls each simulation made independently of the others. When the simulations are run the protocol is copied once for each simulation, so its location in memory can be used to uniquely identify it.
Now that the IterateBCLs class is defined we can create an instance of it and add it to the protocol
bcls = [1500, 1000, 500]
func = IterateBCLs(bcls)
This sets the func instance to be called first at time 20,000ms
and then every 10,000ms until its been run len(bcls)+1 (=4) number of
times
proto.setRunDuring(func, firstRun=10_000, runEvery=10_000, numruns=len(func)+1)
Run the simulation¶
sim_runner = pylqt.RunSim()
sim_runner.setSims(proto)
sim_runner.run()
sim_runner.wait()
Plots!¶
[trace], [meas] = pylqt.DataReader.readAsDataFrame(proto.datadir)
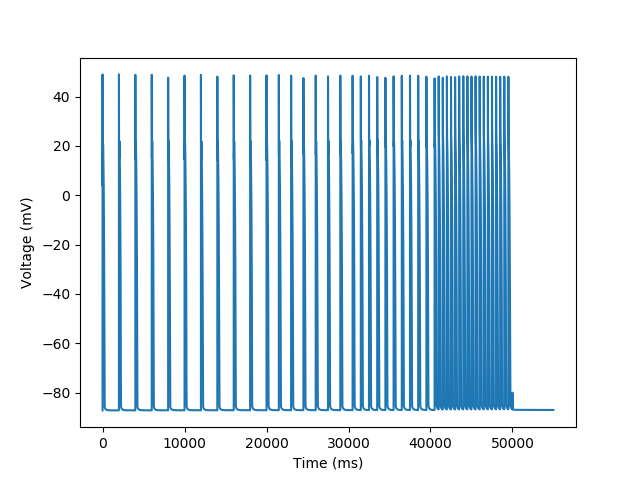
A plot of the voltage over time will show the pacing frequency and we will follow it up with the measured cycle length
plt.figure()
plt.plot('t', 'vOld', data=trace)
plt.xlabel('Time (ms)')
_ = plt.ylabel('Voltage (mV)')
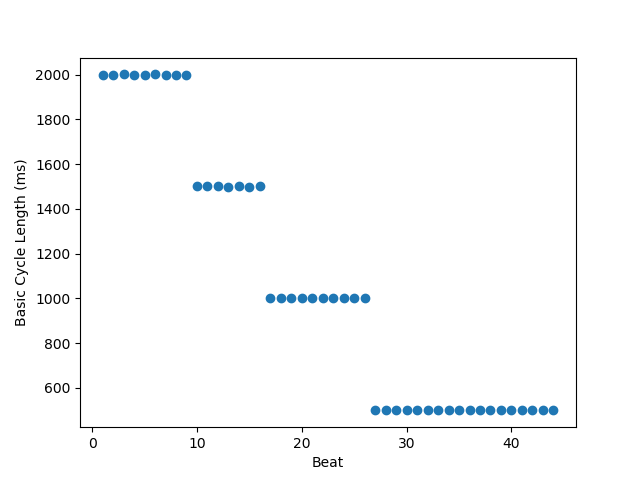
beat = np.arange(meas.shape[0])
plt.figure('Peak Sodium Current vs Beat')
plt.scatter(beat, meas[('vOld', 'cl')])
plt.ylabel('Basic Cycle Length (ms)')
_ = plt.xlabel('Beat')
Total running time of the script: ( 0 minutes 1.291 seconds)