Note
Click here to download the full example code
Parameter Sensitivity Analysis¶
This example illustrates how to conduct parameter sensitivity analysis. The purpose of conducing this kind of analysis is to determine the impact of one (or more) cell parameter, such as sodium ion channel conductance, on one (or more) measurable output, such as upstroke velocity. For this example we will stick to one cell parameter and one output, however the technique is not changed when preforming this analysis for many inputs possible outputs, except that a larger number of simulations may be required. The approach of this analysis can be divided into 3 parts:
Generate a population of cells. The cells in this population are not all the same and instead vary randomly with respect to the parameters of interest. Typically they are generated from a normal or lognormal distribution, who’s spread is tuned to cover a reasonable range of possible inputs. The definition of a reasonable range depends on the purpose of the analysis, for example if the goal is to understand the parameter’s effect in healthy cells, then the population generated should correspond to a range of plausible values for healthy cells.
Run a simulation for each cell in the generated population and measure the resulting outputs.
Conduct a linear regression using the parameters as predictors and the measured output as the response. This creates a linear model who’s coefficients indicated the effect of a change in a parameter to a change in response.
For this example we will specifically be looking at the effect of sodium channel conductance on upstroke velocity.
import PyLongQt as pylqt
import pandas as pd
Setup the Simulation & Configure the population¶
The initial setup is the same as before
proto = pylqt.protoMap['Current Clamp Protocol']()
proto.setCellByName('Canine Ventricular (Hund-Rudy 2009)')
proto.bcl = 800
proto.tMax = 50_000
proto.numstims = 200
proto.meastime = 1_000
proto.writetime = 51_000 #We do not need to write any traces!
proto.cell.variableSelection = set()
proto.measureMgr.percrepol = 90
proto.measureMgr.addMeasure('vOld', {'peak', 'maxderiv'})
The new addition beyond the ‘simple current clamp’ example is the use
of Protocol.pvars to modify the sodium channel conductance for
each simulation.
To get a list of the possible parameters use Cell.pars
list(proto.cell.pars.keys())[:5]
Out:
['AGeo', 'Rcg', 'FDAY', 'InacaFactorbulk', 'Ito2Factor']
Parameter names that end in Factor scale the parameter, so that 1 is the default value for all factors, and their reasonable range is 0 to infinity.
The factor we need is InaFactor, and to set up the pvars entry we need to specify the distribution that will be used to generate the values we will use lognormal, so we will also need to specify the mean and standard deviation of that distribution.
distribution = proto.pvars.Distribution.lognormal
mean = 0
st_dev = 0.2
proto.pvars['InaFactor'] = proto.pvars.IonChanParam(distribution, mean, st_dev)
proto.pvars['InaFactor']
Out:
<TIonChanParam: Lognormal Distribution (Mean: 0.000000 Standard Deviation: 0.200000)>
Finally we need to specify the number of simulations to generate from the popultion. We will generate 20 in this case, although 300-600 is a more typical number.
proto.numtrials = 20
As an aside, the specific values for each trial are stored in
Protocol.pvars and can be viewed (or modified!), below are the
first 5 values
proto.pvars['InaFactor'].trials[:5]
Out:
[1.534903253124452, 0.7789928077395771, 1.0263547171177279, 1.2129733201298847, 0.9115086295274883]
Run the simulations¶
The simulation can be run using the exact same syntax as before.
RunSim automatically detects the number of trails to be run and
handles them concurrently.
sim_runner = pylqt.RunSim()
sim_runner.setSims(proto)
sim_runner.run()
sim_runner.wait()
Read the data & fit linear regression model¶
Reading the data is preformed similarly, however the lists returned now have 20 elements instead of 1, with each DataFrame being for one simulation.
traces, meases = pylqt.DataReader.readAsDataFrame(proto.datadir)
For upstroke velocity we are interested in the final action potential for each simulation rather than all the values, so we will compile that into its own DataFrame first
last_meas = pd.DataFrame([
meas
.loc[meas.index[-1]]
.rename(i)
for i,meas in enumerate(meases)])
A detailed explanation for those unfamiliar with python classes
To unpack a little about how this line of code works, it uses a generator expression […] to generate a list of series which pandas then compiles back into a dataframe. More specifically,
for i,meas in enumerate(meases)gets each individual measure (which we name meas) dataframe from the meases list and enumerates them, 1…20, which we name i. Then, we get the last row from each meas, usingmeas.loc[meas.index[-1]]. Finally, we need to rename thePandas.Series(the row from the dataframe) so that it will have the correct index when they are all compiled into a new dataframe (last_meas), without this last step, all of the rows in the last_meas dataframe would have the same index.
Next we would like to have a copy of the parameter values used in the simulations, which we will get in the same manner as shown earlier.
parameters = pd.Series(proto.pvars['InaFactor'].trials, name='InaFactor')
Now that we have all the data we need we can run a linear regression using the statsmodels pacakge
import statsmodels.api as sm
import seaborn as sns
model = sm.OLS(last_meas[('vOld', 'peak')], sm.add_constant(parameters))
res = model.fit()
print(res.summary())
#%
# The `res` object contains the results of the model fit as well as model
# evaluation criteria, and can be used to determine how much of a change
# in INaFactor will result in a change in upstroke velocity
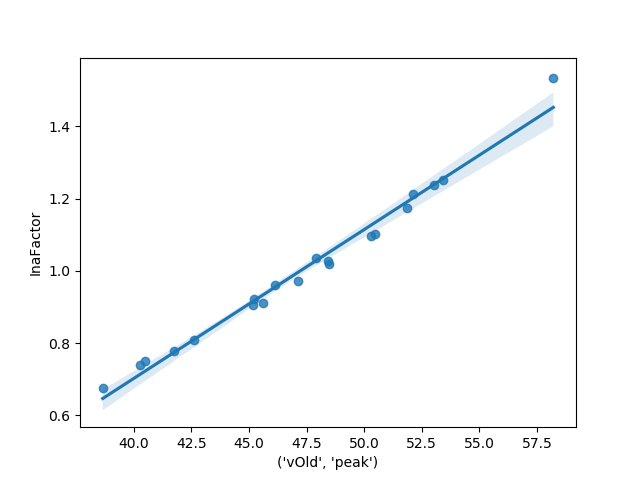
# Finally, as this is a simple linear regression we can generate a plot
# of the points as well as our fit model to examine how strong the model
# looks
_ = sns.regplot(last_meas[('vOld', 'peak')], parameters)
Out:
C:\Users\grat05\AppData\Local\Programs\Python\Python37\lib\importlib\_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject
return f(*args, **kwds)
OLS Regression Results
==============================================================================
Dep. Variable: ('vOld', 'peak') R-squared: 0.983
Model: OLS Adj. R-squared: 0.982
Method: Least Squares F-statistic: 1059.
Date: Mon, 04 Oct 2021 Prob (F-statistic): 1.91e-17
Time: 16:27:48 Log-Likelihood: -19.488
No. Observations: 20 AIC: 42.98
Df Residuals: 18 BIC: 44.97
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 23.3579 0.753 31.019 0.000 21.776 24.940
InaFactor 23.8657 0.734 32.535 0.000 22.325 25.407
==============================================================================
Omnibus: 5.869 Durbin-Watson: 1.368
Prob(Omnibus): 0.053 Jarque-Bera (JB): 3.579
Skew: -0.962 Prob(JB): 0.167
Kurtosis: 3.769 Cond. No. 9.87
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
This model naturally fits quite well. In other scenarios it may be necessary preform a more complete linear regression with model diagnostics, etc to insure model quality. There are many tutorials on linear regression that cover the appropriate steps for running a regression.
Total running time of the script: ( 0 minutes 4.684 seconds)